Protein-DNA interactions dictate gene expression and replication. Although several techniques are able to detect the location of an interaction, few are able to measure its strength, a critical determinant of DNA accessibility. Our lab initially developed the ‘unzipping mapper’ technique as a powerful and versatile DNA unzipping method to measure protein-DNA interactions (Koch et al., BJ, 2002; Koch and Wang, PRL, 2003; Shundrovsky et al., NSMB, 2006; Hall et al., NSMB, 2009). Using an optical trap, we mechanically separate a double-stranded DNA (dsDNA) with bound proteins into two single strands. As the unzipping fork reaches a bound protein, the unzipping force increases dramatically above the DNA unzipping baseline. The interaction of the fork with the bound protein detects positions to near single-base-pair precision, while also providing a quantitative measure of their strengths. Our recent additions of ‘unzipping tracker’ and ‘unzipping staller’ (Le et al, Cell, 2018) expand the utility of our pioneering unzipping technique.
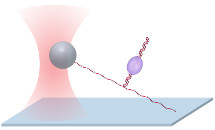 Unzipping Mapper Unzipping MapperOur ‘unzipping mapper’ disrupts each interaction sequentially within a complex along DNA, creating a detailed map of locations and strengths of multiple interactions within a large protein-DNA complex to near base-pair resolution (Shundrovsky et al., NSMB, 2006, Hall et al., NSMB, 2009). |
The DNA “unzipping tracker” assay monitors, in real time, translocation of a translocase (Le et al., Cell, 2018) or a helicase (Johnson et al., 2007; Sun et al., 2011) without the need to tag or anchor the motor protein. |
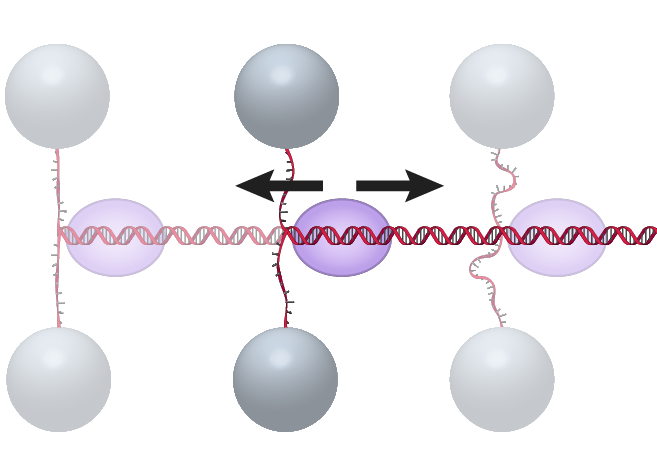 Unzipping Staller Unzipping StallerThe DNA “unzipping staller” assay uses a DNA fork as a barrier to stall a DNA translocase moving toward the fork (Le et al, Cell, 2018). The DNA is unzipped towards the translocated motor, but before reaching the motor, the unzipping is paused by holding the unzipped single strand DNA at a fixed extension. As the motor reaches the stalled unzipping fork it will begin to rezip the DNA and the force in the remaining single strand tether will increase until the motor protein stalls, dissociates, or backtracks. |
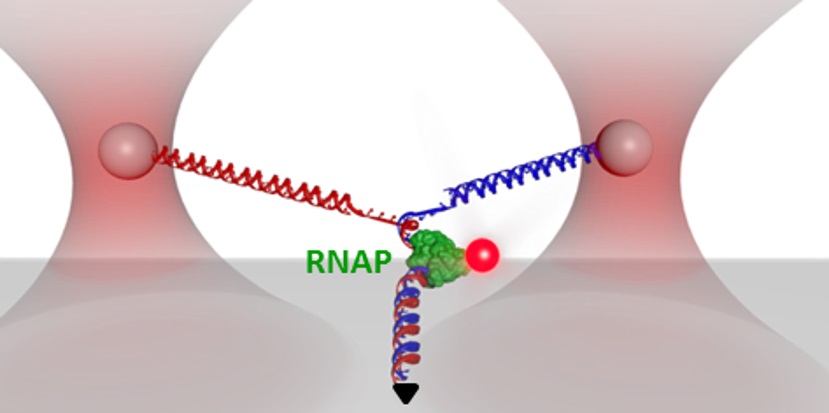 Y Structure Y StructureCurrent implementations of optical trapping usually involve only stretching, unzipping, or twisting DNA along one dimension. To expand the capabilities of optical trapping for more complex measurements we developed a multidimensional technique that combines all of these manipulations in a single experiment. Our three-branch DNA construct, termed a “Y structure’ mimics the replication fork and allows DNA unzipping under supercoiling, as well as fluorescence visualization of bound proteins, on the DNA (Inman et al., Nano Letters, 2014). This multidimensional assay allows precise, real-time tracking of multiple configurational changes. |
