DNA topology is constantly being restructured by the action of motor proteins. During transcription, RNAP over twists DNA in the front (downstream) and under twists DNA behind (upstream), as described by the twin‐supercoiled domain model. This transcription‐generated torsion is present genome‐wide and can significantly facilitate or inhibit gene expression.
Historically, the most prevalent approach to study DNA topology changes has been the use of 2D gels to measure one aspect of torsion – the degree of supercoiling. However, this cannot measure torque – how difficult it is to twist DNA – the key parameter that characterizes torsion and determines whether DNA is denatured and whether a bound protein is evicted.
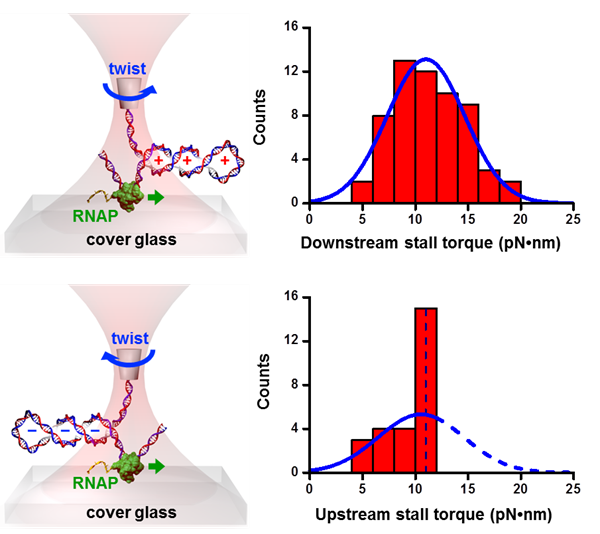 RNAP RNAPWe developed the angular optical trap (AOT) for direct torque measurements (Deufel et al., Nature Methods, 2007) and using the AOT, we made the first direct torque measurement of any DNA‐based motor by showing that E. coli RNAP generates ~ 11 pN∙nm of torque (Ma et al, Science, 2013). |
 GreB GreBBecause we observed that RNAP backtracking limits RNAP torque generation capacity, we considered whether transcription factors such as GreB, which rescues backtracking, could increase torque generation capacity. We found that GreB greatly limits RNAP backtracking and increases torque capacity by 65% (to 18.5 pN∙nm) and through de novo sophisticated statistical mechanics analysis of the stalling trajectories and determined the kinetic rates of backtracking and GreB rescue (Ma et al, bioRxiv, 2018). |
 Nucleosome under torsion Nucleosome under torsionRNAP must transcribe through nucleosomes which present formidable barriers to transcription. We found that torque could regulate histone exchange during transcription and replication (Sheinin et al., Nature Communications, 2013). |
